User Tools
Table of Contents
Limits with the Higgs Combined Tool
The Combined Tool by the Higgs Combination Group provides the combine command which runs different statistical methods with RooStats for some input, e.g. a text file (datacard) containing information on the systematic uncertainties, data and processes, like the example below. It is extensively discussed here:
- TWiki documentation: https://twiki.cern.ch/twiki/bin/viewauth/CMS/SWGuideHiggsAnalysisCombinedLimit
- GitBooks documentation: https://cms-hcomb.gitbooks.io/combine/content/
- Hypernews forum: https://hypernews.cern.ch/HyperNews/CMS/get/higgs-combination
These are useful tutorials on the Combine Tool from a 2016 CERN workshop (slides plus video). It may be interesting to do the exercises on SWAN. Otherwise, take a look at these examples on GitHub.
Many other examples can be found on this TWiki page for the CMSDAS school.
Description of the statistical methods at CMS:
- CMS Collaboration (2012), “Observation of a new boson with a mass near 125 GeV”, CMS-PAS-HIG-2012/020.
- Cowan et. al (2013), “Asymptotic formulae for likelihood-based tests of new physics”,arXiv:1007.1727.
- This four-part lecture by Glen Cowan covers a more theoretical overview of statistical methods used in Particle Physics.
CombineHarvester is a tool to manage datacards: its GitHub repository and the full documentation.
Quick setup
Latest instructions to get combine can be found in the manual and on the TWiki. In summary: Get CMSSW for the Combined Tool:
export SCRAM_ARCH=slc6_amd64_gcc530 cmsrel CMSSW_8_1_0 cd CMSSW_8_1_0/src cmsenv
(combine in CMSSW_7_4_7 is outdated.) Get Combined Tool:
git clone https://github.com/cms-analysis/HiggsAnalysis-CombinedLimit.git HiggsAnalysis/CombinedLimit cd HiggsAnalysis/CombinedLimit git fetch origin git checkout v7.0.8 scramv1 b clean; scramv1 b
With every new session in PSI Tier3, set up the environment with
source $VO_CMS_SW_DIR/cmsset_default.sh export SCRAM_ARCH=slc6_amd64_gcc530 cd ~/phase2/CMSSW_8_0_1/src/ cmsenv
Simple example: Running the combine tool
To run the tool you need to make a datacard containing information on the nuisance parameters of the signal and background processes. It will look like this example from the official tutorial:
- HHdatacard.txt
# Simple counting experiment, with one signal and a few background processes # Simplified version H->WW analysis for mH = 160 GeV imax 1 number of channels jmax 3 number of backgrounds kmax 4 number of nuisance parameters (sources of systematical uncertainties) -------------------------------------------------------------------------------------- # one channel, 0 observed events bin 1 observation 0 -------------------------------------------------------------------------------------- bin 1 1 1 1 process ggH qqWW ggWW others process 0 1 2 3 rate 1.47 0.63 0.06 0.22 -------------------------------------------------------------------------------------- lumi lnN 1.11 - 1.11 - luminosity; lnN = lognormal xs_ggH lnN 1.16 - - - gg->H cross section + signal efficiency xs_ggWW lnN - - 1.50 - gg->WW cross section bg_others lnN - - - 1.30 30% uncertainty on the rest of the backgrounds
To get the expected 95% upper limit on the signal strength with the asymptotic CLs method run
combine -M Asymptotic -m 160 -n .HH HHdatacard.txt
which will prompt the upper limits r and output a ROOT file higgsCombine.HH.Asymptotic.mH160.root with all the results. 50.0% stands for the expected median, 16%-84% for the +/- one standard deviation 𝜎 and 2.5-97.5% for two. The option -m <M> is only used as a label of the particle mass M in the tree and file naming as you might vary this parameter at a later stage. Option -n <name> allows for a custom output name.
Creating datacards
If you have many different processes, channels and nuisance parameters, it can be hard to keep track of the datacards manually. CombineHarvester is a tool that allows you to quickly create one or more datacards with one python or C++ script. For instructions and examples, please take a look at the GitHub repository and the full documentation.
In the simple example above all nuisance parameters are assumed to be log-normally distributed lnN. The tutorial lists other options, like the Gamma (gmN) and log-uniform (lnU) distributions, and the motivations for them.
Some systematic uncertainties however, may influence the final selections and variable shapes. Say you are using a tau object to calculate the final discriminating variable like the invariant mass. Then the analysis needs to be run again with an one standard deviation up and down shift of the tau energie scale (TES), propagated to this variable. These shapes (histograms), then, can be in included in the datacard as a nuissance parameter by using shape as distribution. The path to the ROOT file containing the histograms of each process plus the histograms where the relevant shift has been applied, needs to be included in the datacard.
Shapes can also be probability density functions (PDFs), which can be parametrized and is stored as a RooAbsPdf in a RooWorkspace in a ROOT file.
Read more here.
Note there are some general datacard guidelines and naming schemes you should follow, found here.
Other methods and options
The combine tool has many more methods and options (type combine --help).
If you are interested in the expected significance, use MaxLikelihoodFit (FitDiagnositcs in CMSSW_8_1_0). This can method also can be used to extract the observed cross section.
If you are interested in only an approximation of the expected number of signal events (yield) with the maximum likelihood fit, use ProfileLikelihood. For the upper limits however, you always should use Asymptotic instead.
For analysis with blinded data, you can use the option -t -1, which will create a Asimov dataset with the background Monte Carlo samples. Otherwise you can simulate the dataset with toys using -t <N> for N toys.
Analysis of the systematic uncertainties
These slides of the aforementioned workshop explain how to separate the systematical from the statistical uncertainties, and how to analyse the impact and pull of each parameter. It makes use of the combine method MultiDimFit and scripts provided by the CombineHarvester tool.
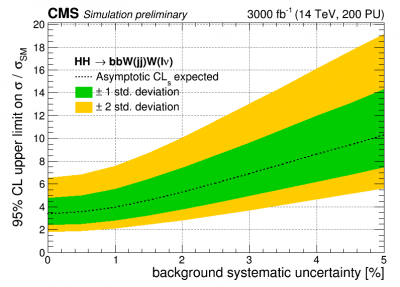
Brazilian plot
To make a Brazilian plot, you can make a simple python script that automatically creates the datacards for different values of some parameter like the invariant mass. This example shows scan for the background uncertainty. Guidelines for a CMS-style limit plot can be found here and here.
Other
- BibTeX reference to asymptotic CLs method:
@article{CLs, author = {Cowan, Glen and Cranmer, Kyle and Gross, Eilam and Vitells, Ofer}, title = {Asymptotic formulae for likelihood-based tests of new physics}, journal = {The European Physical Journal C}, volume = {71}, number = {2}, issn = {1434-6052}, year = {2011}, month = {Feb}, doi = {10.1140/epjc/s10052-011-1554-0}, eprint = {1007.1727}, archivePrefix = {arXiv}, primaryClass = {hep-ph}, SLACcitation = {%%CITATION = arXiv:hep-ph/1007.1727;%%}, }

